Photo created by me on canva
First of all, dealing with data in a PDF file can be a nightmare, not to mention those containing a table or even tables. Most often, those tables contain the important data we want to analyse. Although there are different Python packages for extracting data from a PDF, I prefer pdfplumber because of the ease of extracting tabular data.
PDFplumber extracts tables, but not in a way we might intend them to look. It extracts tables inside a list, more like a list of lists, where each row of data is in a list inside a list containing all rows.
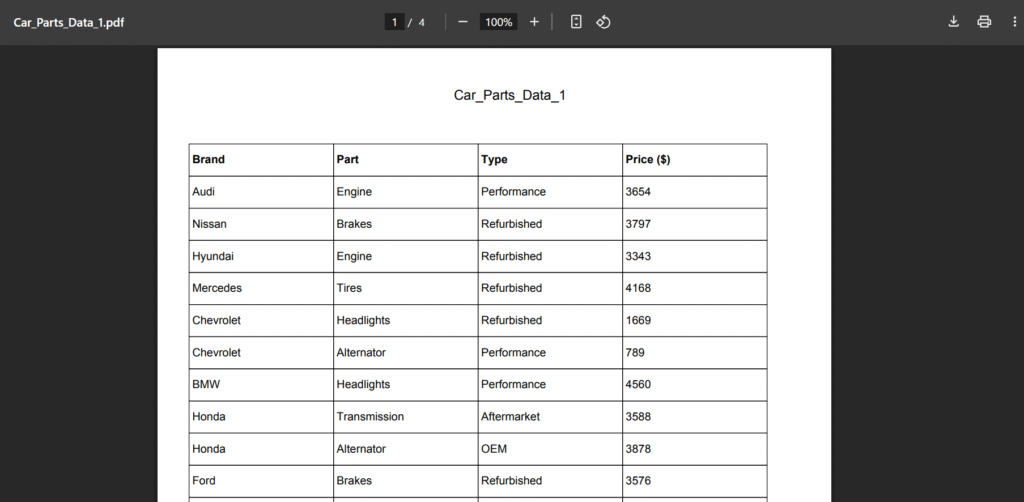
Let’s take the PDF image above, I randomly generated the PDF using ChatGPT for this extraction. Note that we might want to extract the data exactly as seen, but pdfplumber will give us a list of lists:
import pdfplumber
#Using context manager
with pdfplumber.open(r"pdfs\Car_Parts_Data_1.pdf") as pdfs:
page = pdfs.pages[0]
page_table = page.extract_table()
print(page_table)#Output
[['Brand', 'Part', 'Type', 'Price ($)'], ['Audi', 'Engine', 'Performance', '3654'], ['Nissan', 'Brakes', 'Refurbished', '3797'], ['Hyundai', 'Engine', 'Refurbished', '3343'], ['Mercedes', 'Tires', 'Refurbished', '4168'], ['Chevrolet', 'Headlights', 'Refurbished', '1669'], ['Chevrolet', 'Alternator', 'Performance', '789'], ['BMW', 'Headlights', 'Performance', '4560'], ['Honda', 'Transmission', 'Aftermarket', '3588'], ['Honda', 'Alternator', 'OEM', '3878'], ['Ford', 'Brakes', 'Refurbished', '3576'], ...]In some cases, this output might be sufficient for us to work with, but in most cases, we might need our data to be in a dataframe, just as in the screenshot above. This is where my code comes in.
Dependencies
import pdfplumber
import pandas as pd
import osWe would start by importing this two packages
- pdfplumber: This is the heavy lifter when it comes to parsing text and tables from PDFs.
- pandas: For organizing our table data into structured, easy-to-work-with DataFrames.
- os: This is not really necessary, nor was it included in the code logic, but it may be handy for handling file paths in more complex workflows, which you will see in an example.
If you don’t have this packages simply type on your terminal:
pip install pdfplumber pandasThe ReadPdf_Table class
class ReadPdf_Table():
def __init__(self, filename, table_column_num = 20):
self.filename = filename
self.table_column_num = table_column_num
self.table_dict = {} This is the class that encapsulates all the functionality of our code, which includes loading a PDF, extracting tables, grouping them, and converting them into a pandas Dataframe (which is best for data analysis) upon initialization.
- filename: This is the path to the PDF file
- table_column_num: This is a configurable maximum number of columns to be considered when grouping a table. You can set it to any number of columns, but my default is 20 (simple tables are usually less)
- table_dict: This will store grouped tables based on their column count.
The table_dict is very necessary because some PDFs contain more than one table with different numbers of columns
Loading the Table into a Dataframe
def loadTable(self):
"""Public method: Extracts and returns PDF tables as DataFrames."""
self.table_dict = self._pdf_toDict()
self.dataframes = self._convert_toTable(self.table_dict)
return self.dataframesThis is a public method of the ReadPdf_Table class; it is the only method accessible outside of the scope of the class, and its function is to return the converted table as a Dataframe.
The sequence of what it does:
- Calls a private method
_pdf_toDict()to scan all pages of the PDF and store extracted tables inself.table_dict. - Passes the resulting dictionary to
_convert_toTable()to convert the raw table data into pandas DataFrames. - Returns a list of cleaned DataFrames ready for analysis.
This separation is crucial, extracting raw tables first, then converting them, is clean and allows for better debugging or customization (like extracting more than one PDF in a folder having the same table structure and appending them as one Dataframe).
The next two private methods, _pdf_toDict() and _convert_toTable() are the next to be discussed, they are the two major function that makes this extraction possible.
Step 1: Extract Tables Into a Dictionary
I will break this part down for better understanding
def _pdf_toDict(self):
"""Internal method: Extracts tables from PDF into a dictionary."""
with pdfplumber.open(self.filename) as pdf:
num_pages = len(pdf.pages)The first step in this method is to use a Contest Manager to open the PDF with pdfplumber.
Most often, PDFs might be more than one page, and we need to account for all the pages that the table contains. Account statements, e-commerce product documents, and accounting documents are examples of PDF documents that have tables that can span across the entire document.
As seen in the code above, we can get the number of pages in a PDF after reading it using len(pdf.pages).
for page_num in range(num_pages):
page = pdf.pages[page_num]
page_table = page.extract_table()At this point, the number of pages is automatically handled for us, whether it is a 5-page PDF or a 1000-page PDF, it will give us the accurate page number.
Now that we have the total number of pages, we have to loop through each page to extract all the data in the table. If a PDF contains two sets of tables, say a three-column table and a four-column table, it will extract them into a list of three index lists and four index lists.
Example:
- A list of three index lists – [[“A”, “B”, “C”], [“D”, “E”, “F”]].
- A list of four index lists – [[“A”, “B”, “C”, “D”], [“E”, “F”, “G”, “H”]].
page_table will look like – [[“A”, “B”, “C”], [“D”, “E”, “F”], [“A”, “B”, “C”, “D”], [“E”, “F”, “G”, “H”]]
for table_column in page_table:
for _ in range(self.table_column_num):
if len(table_column) == _:
table_column_name = f'table_columns_{str(_)}'
if table_column_name not in self.table_dict.keys():
self.table_dict[table_column_name] = [table_column]
else:
self.table_dict[table_column_name] += [table_column]
This is the final section of the _pdf_toDict() method. This part is to separate the tables into appropriate columns, provided that the PDF has tables with different column numbers.
Take for example, [[“A”, “B”, “C”], [“D”, “E”, “F”]] and [[“A”, “B”, “C”, “D”], [“E”, “F”, “G”, “H”]] are three column and four column table respectively in a PDF, how will the above code run?
- for table_colum in page_table: – This part takes the total rows (lists) of all tables and runs them based on their index, e.g.
[["A", "B", "C"], ["D", "E", "F"], ["A", "B", "C", "D"], ["E", "F", "G", "H"]]where["A", "B", "C"]is index 0, ["D", "E", "F"]["A", "B", "C", "D"]is index 2 and so on. - for _ in range(self.table_column_num): – This is the range of total allowable column number as provided upon initialisation, which we gave a value of 20 allowable columns.
- if len(table_column) == _: – This is where the table separation is done based on how many columns it has. Let’s take the table_column of index 0, which is
["A", "B", "C"]. This row has a length of 3, which means it has 3 columns. We give it a table_column_name based on the number of columns it has, which is shown by the formatted string value f’table_columns_{str(_)}’. This gives ‘table_columns_3’ if the column length is 3 - if table_column_name not in self.table_dict.keys(): – This part is responsible for adding data of the same column number to a dictionary, if not been added.
- self.table_dict[table_column_name] = [table_column]: – Adding a new column key to the dictionary.
- self.table_dict[table_column_name] += [table_column]: – Appending a new row (list) to an already existing column with the same number of columns.
return self.table_dictFinally, we return the table dictionary – self.table_dict.
Below is the complete _pdf_toDict() method:
def _pdf_toDict(self):
"""Internal method: Extracts tables from PDF into a dictionary."""
with pdfplumber.open(self.filename) as pdf:
num_pages = len(pdf.pages)
for page_num in range(num_pages):
page = pdf.pages[page_num]
page_table = page.extract_table()
for table_colum in page_table:
for _ in range(self.table_column_num):
if len(table_colum) == _:
table_column_name = f'table_colums_{str(_)}'
if table_column_name not in self.table_dict.keys():
self.table_dict[table_column_name] = [table_colum]
else:
self.table_dict[table_column_name] += [table_colum]
return self.table_dictStep 2: Convert to Cleaned DataFrames
This part is a bit straightforward. We convert our dictionary to a Dataframe.
def _convert_toTable(self, dictionary):
"""Internal method: Converts dictionary to cleaned DataFrames."""
self.dictionary = dictionary
self.dataframes = []First, we define our method variables. _convert_toTable(self, dictionary) takes a dictionary parameter as its argument. This dictionary will be the dictionary returned by the _pdf_toDict() method. We also need to define the dataframe as a list so we can extract any dataframe of choice based on the number of columns they have.
for key, values in self.dictionary.items():
column_header = values[0]
content = []
for value in values[1:]:
if value == column_header:
pass
else:
content.append(value)
dataframe = pd.DataFrame(content, columns=column_header)
clean_df = dataframe.dropna(how='all')
self.dataframes.append(clean_df)In this section, we loop into the dictionary where the keys are the number of columns the table has, and the values are the row data.
As with most data, the first row is usually the header, so it was specified in the second line of code – column_header = values[0]
The third line, content = [] is for saving the remaining values as the table data after the table header
#Example dictionary
table_dict = {
'table_columns_3': [['Brand', 'Part', 'Type', 'Price ($)'], ['Audi', 'Engine', 'Performance', '3654'], ['Nissan', 'Brakes', 'Refurbished', '3797']]
'table_columns_4': [['Brand', 'Part', 'Type', 'Country', 'Price ($)'], ['Mercedes', 'Tires', 'Refurbished', 'Germany', '4168'], ['Chevrolet', 'Headlights', 'Refurbished', 'USA,'1669'], ['Toyota', 'Alternator', 'Performance', 'Japan', '789']]
}
Say the above dictionary is passed to the _convert_toTable(self, dictionary) method, for key ‘table_columns_3’, the list [‘Brand’, ‘Part’, ‘Type’, ‘Price ($)’], is values[0], which will be saved as the column_header. The remaining lists [‘Audi’, ‘Engine’, ‘Performance’, ‘3654’] and [‘Nissan’, ‘Brakes’, ‘Refurbished’, ‘3797’] will be appended to the content list.
if value == column_header: pass – This part takes care of duplicate headers, which usually occur when a table spans across multiple PDF pages, so whenever it comes across the same header, it skips it
- dataframe = pd.DataFrame(content, columns=column_header): – Converts the dictionary key and values to a Dataframe
- clean_df = dataframe.dropna(how=’all’): – Drop any row with no data
- self.dataframes.append(clean_df): – Append the new dataframe to the dataframes list
return self.dataframes And finally, we return self.dataframes
Below is the complete _convert_toTable() method:
def _convert_toTable(self, dictionary):
"""Internal method: Converts dictionary to cleaned DataFrames."""
self.dictionary = dictionary
self.dataframes = []
for key, values in self.dictionary.items():
column_header = values[0]
content = []
for value in values[1:]:
if value == column_header:
pass
else:
content.append(value)
dataframe = pd.DataFrame(content, columns=column_header)
return self.dataframes Wrap-up
At this point, we have a working code that can read and extract tables to a Dataframe from a PDF. You can see the full code on my GitHub account.
reader = ReadPdf_Table(r"pdfs\Car_Parts_Data_1.pdf")
dataframes = reader.loadTable()
print(dataframes[0])#Output
Brand Part Type Price ($)
0 Audi Engine Performance 3654
1 Nissan Brakes Refurbished 3797
2 Hyundai Engine Refurbished 3343
3 Mercedes Tires Refurbished 4168
4 Chevrolet Headlights Refurbished 1669
.. ... ... ... ...
95 Chevrolet Transmission Aftermarket 1561
96 Hyundai Transmission Performance 4924
97 Kia Alternator Refurbished 617
98 Honda Headlights OEM 2045
99 Kia Radiator Refurbished 289
[100 rows x 4 columns]If there is more than one dataframe, you can access them using the index, You can also use len(dataframes) to know how many dataframes are in the dataframes list
Tweaking
Remember, at the beginning, I said something about using the os module for handling file paths in more complex workflows like extracting more than one PDF in a folder having the same table structure and appending them as one Dataframe. In the code below, I extracted 5 car_parts_data PDFs with 100 rows of data each, making a total of 500 rows of data as a single Dataframe.
folder_path = r"pdfs"
file_paths = [os.path.join(folder_path, file) for file in os.listdir(folder_path) if file.lower().endswith('.pdf')]
all_data = pd.DataFrame()
for path in file_paths:
table = ReadPdf_Table(filename=path)
data = table.loadTable()[0]
all_data = pd.concat([all_data, data], ignore_index=True)
print(all_data)#Output
Brand Part Type Price ($)
0 Audi Engine Performance 3654
1 Nissan Brakes Refurbished 3797
2 Hyundai Engine Refurbished 3343
3 Mercedes Tires Refurbished 4168
4 Chevrolet Headlights Refurbished 1669
.. ... ... ... ...
495 Chevrolet Radiator OEM 4031
496 Kia Radiator Performance 2711
497 Honda Battery Refurbished 110
498 Chevrolet Transmission Performance 1880
499 Kia Alternator Refurbished 3717
[500 rows x 4 columns]Final Thought
This class offers a flexible foundation for PDF table extraction. You could improve it further by:
- Adding error handling for
extract_table()failures. - Including logging or progress output.
- Adding options for treating merged cells or skewed formatting.
Although this code extracts PDFs containing one or more tables but if there are two different tables with similar columns, it might append them as one. This can be separated while using pandas to manipulate and clean up dataframe after extraction.
Note: I am not perfect, you might see a mistake or a better way of doing this same thing, I will be glad if you are willing to share. You can check out some of my project works here.
As it stands, this is a solid starting point for anyone looking to bring tabular data from PDFs into the Python data science ecosystem.